From predicting protein structures to diagnosing diseases from images, AI is transforming biology. At the heart of this transformation are deep learning architectures for biologists—specialized tools designed to help researchers uncover patterns in complex data. Terms like CNNs, RNNs, and transformers might seem intimidating, but they’re easier to understand than they appear.
This guide breaks down these architectures in simple terms, showing how they work and how they’re applied in real-world biology. Whether you’re a researcher, a student, or just curious, this is your starting point to understanding how deep learning is shaping the future of life sciences.
What Are AI, Machine Learning, Neural Networks, and Deep Learning?
Understanding deep learning starts with grasping its foundations: artificial intelligence (AI), machine learning (ML), neural networks, and finally, deep learning. Here’s a simple breakdown to guide you through these interconnected concepts.
1. Artificial Intelligence (AI): Machines That Mimic Human Intelligence
AI is a broad field focused on creating systems that can perform tasks traditionally requiring human intelligence, such as recognizing speech, making decisions, or analyzing data. It’s the overarching term that encompasses all efforts to make machines “think” or act intelligently.
2. Machine Learning (ML): Teaching Machines to Learn from Data
Machine learning is a subset of AI where systems learn patterns from data instead of being explicitly programmed. It’s like teaching a machine to make decisions by showing it examples, rather than writing rules for every situation.
3. Neural Networks: Mimicking the Human Brain
Neural networks are the foundation of machine learning. They’re inspired by the human brain, using layers of interconnected nodes (like neurons) to process data. Each layer extracts specific features, gradually building an understanding of the data.
4. Deep Learning: Advanced Neural Networks for Complex Problems
Deep learning takes neural networks to the next level by adding multiple layers, creating “deep” networks capable of solving highly complex tasks. These layers work together to automatically extract and refine features from raw data.
How These Concepts Connect
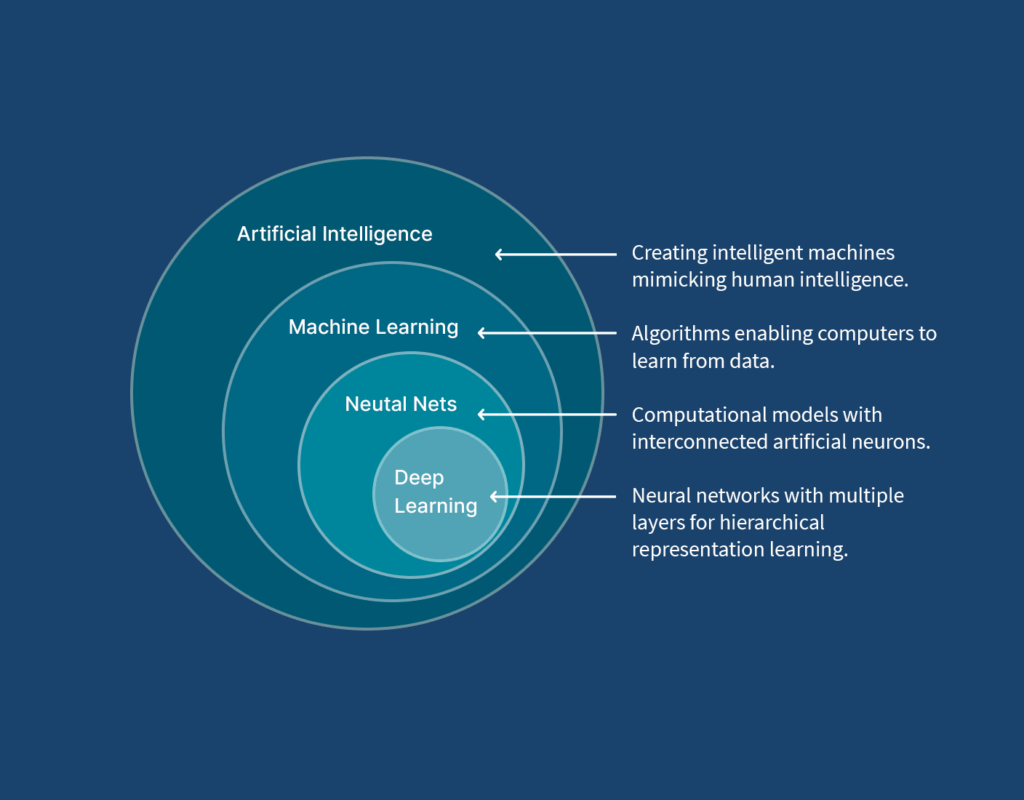
Think of AI as the big picture, machine learning as a tool within AI, neural networks as the structure powering machine learning, and deep learning as the most advanced version of neural networks. Together, they provide powerful tools for solving biological challenges, from uncovering genetic secrets to optimizing lab experiments.
A Simple History of Deep Learning for Biologists
Deep learning has come a long way from its origins in simple neural models to becoming a key tool in modern biology. Its evolution is a story of overcoming technological and conceptual challenges to solve increasingly complex problems.
1. Early Inspiration: Mimicking the Brain
In the 1950s, researchers created the first neural networks, inspired by how the brain processes information. These models aimed to replicate the way neurons communicate and learn, using mathematical equations to simulate brain-like behavior.
- The Perceptron, developed in 1958, was one of the first models capable of recognizing basic patterns. However, it struggled with tasks involving overlapping or non-linear data, such as distinguishing shapes or predicting outcomes.
2. The AI Winter: Stalling Progress
By the 1970s and 1980s, enthusiasm for neural networks waned. Early models lacked the depth and computational power to handle real-world problems, and researchers shifted their focus elsewhere.
- Limited layers in these networks meant they couldn’t process complex biological or visual data. For instance, early models couldn’t analyze DNA sequences or detect patterns in imaging datasets.
- Funding cuts and skepticism about AI’s potential led to a period of stagnation known as the “AI winter.”
3. A Revival Sparked by Technology
In the 2000s, deep learning experienced a resurgence thanks to advancements in three areas:
- Better Learning Algorithms: Techniques like backpropagation allowed neural networks to adjust and improve based on errors.
- Access to Big Data: Digitization of information—from genomic databases to medical imaging archives—provided the fuel for training large models.
- Improved Hardware: Graphics processing units (GPUs) revolutionized how efficiently models could be trained.
- In 2012, a breakthrough came with AlexNet, a deep convolutional neural network that outperformed traditional methods in image recognition tasks. It proved deep learning’s potential for real-world applications, including biological research.
4. Today: Deep Learning’s Role in Biology
Modern deep learning architectures, such as convolutional neural networks (CNNs), transformers, and graph neural networks (GNNs), have unlocked new possibilities in biology. These models can process vast amounts of data and uncover insights faster than traditional methods.
Deep learning has also enabled tasks like mapping gene interactions, predicting drug effects, and simulating ecological networks.
Tools like AlphaFold use deep learning to predict protein structures with unprecedented accuracy, while CNNs help analyze medical images for disease detection.
Core Deep Learning Architectures
Deep learning architectures are the backbone of many AI-powered tools in biology. Each architecture is designed to handle specific types of data and tasks, from analyzing images to processing genetic sequences. Here’s a breakdown of the most important architectures and how they’re used in biological research.
1. Convolutional Neural Networks (CNNs): AI for Images
Imagine you’re trying to find a single diseased cell in a microscope image with thousands of healthy ones. That’s where CNNs excel. Convolutional Neural Networks are designed to analyze images by breaking them into smaller parts and detecting patterns like edges, shapes, or textures.
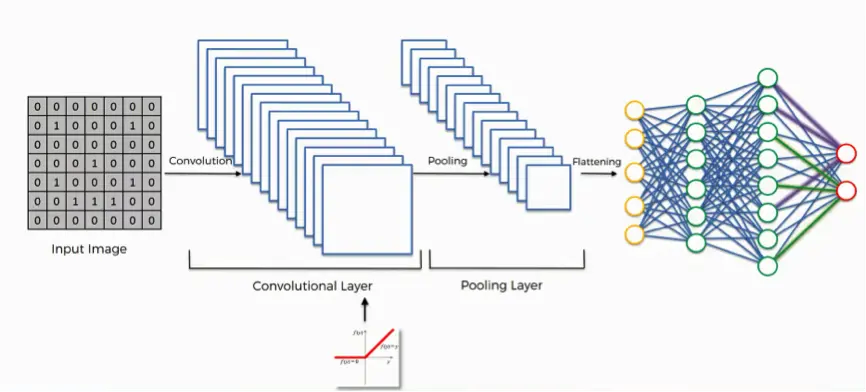
- How CNNs Work: CNNs apply filters across an image to identify basic patterns in the first layers, such as straight lines or curves. As the data moves through deeper layers, the network combines these patterns into more complex structures, such as cells, tissues, or even entire organisms. This layered approach makes CNNs extremely effective for image analysis.
- Biology in Action: CNNs are widely used in medical imaging. For instance, they help pathologists detect cancerous cells in tissue samples by highlighting abnormalities invisible to the naked eye. In plant biology, CNNs can analyze leaf images to diagnose diseases or nutrient deficiencies, helping farmers manage crops more effectively.
2. Recurrent Neural Networks (RNNs): AI for Sequences
Biological processes often occur in sequences. Think of the progression of gene expression over time or the sequence of amino acids in a protein. RNNs are built to analyze data where the order of information matters. Unlike traditional models, RNNs remember past inputs as they process new data, making them ideal for sequential tasks.
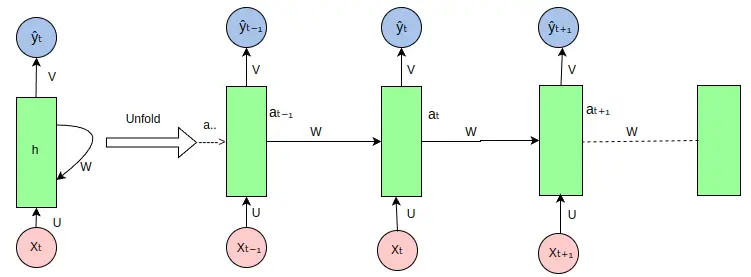
- How RNNs Work: RNNs use loops within their architecture to retain information from earlier steps. For example, when analyzing a DNA sequence, an RNN considers not just the current nucleotide but also the ones before it. This context-aware processing allows the model to identify patterns that span across the sequence.
- Biology in Action: In proteomics, RNNs predict how proteins fold by analyzing the sequence of amino acids. They’re also used to study population dynamics over time, helping ecologists understand how species interact in changing environments.
3. Generative Adversarial Networks (GANs): AI for Synthetic Data
Imagine a system that can create realistic biological data from scratch. That’s the power of GANs. These networks consist of two parts: a generator that creates synthetic data and a discriminator that evaluates its quality. Through this adversarial process, GANs learn to produce increasingly realistic outputs.
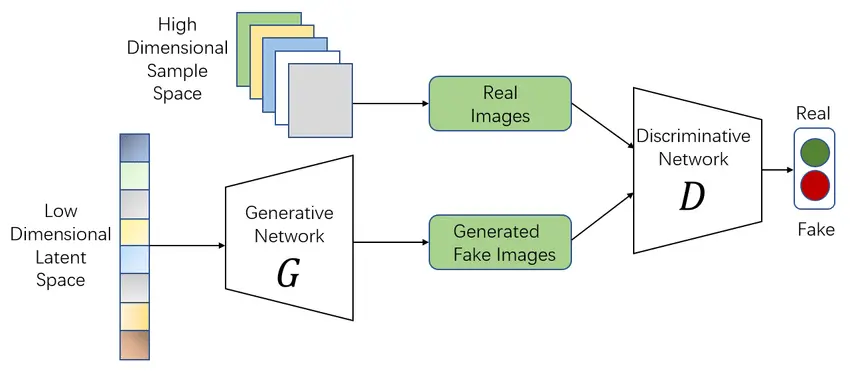
Biology in Action: GANs are invaluable when real-world data is scarce. For example, they can generate synthetic microscopy images to train other AI models without the need for extensive manual data collection. In drug discovery, GANs create new molecular structures that are tested computationally before moving to the lab.
How GANs Work: The generator starts by producing random outputs, like synthetic cell images. The discriminator evaluates these against real data and provides feedback to the generator. Over time, the generator improves until its outputs are indistinguishable from real data.
Read more: Generative Models in Drug Design
4. Transformers: AI for Contextual Understanding
Biological data, such as genetic sequences or protein interactions, often involves relationships across elements. Transformers are particularly good at identifying these relationships because they use a mechanism called self-attention. This allows the model to focus on the most important parts of the data while considering the entire context.
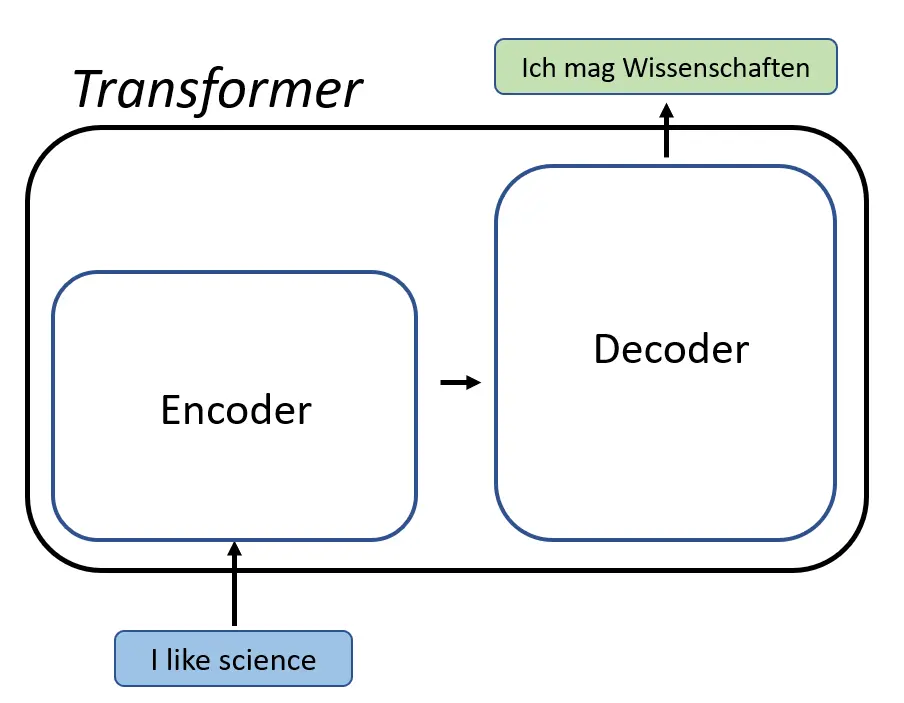
- How Transformers Work: Unlike RNNs, which process sequences step by step, transformers analyze the entire sequence at once. For example, in a DNA sequence, the model can simultaneously evaluate how one nucleotide relates to others, no matter how far apart they are.
- Biology in Action: Transformers are revolutionizing fields like genomics. For instance, they help researchers predict protein structures by analyzing how amino acids interact within a sequence. They’re also used in drug discovery to design molecules tailored to specific targets.
Read More: Transformers in Drug Design
5. Encoder-Decoder Architectures: Transforming Data
Some tasks require converting data from one form to another, such as translating genetic sequences into functional annotations. Encoder-decoder architectures are specifically designed for such transformations. They first compress the input data into a simplified form (encoding) and then reconstruct it into the desired output format (decoding).
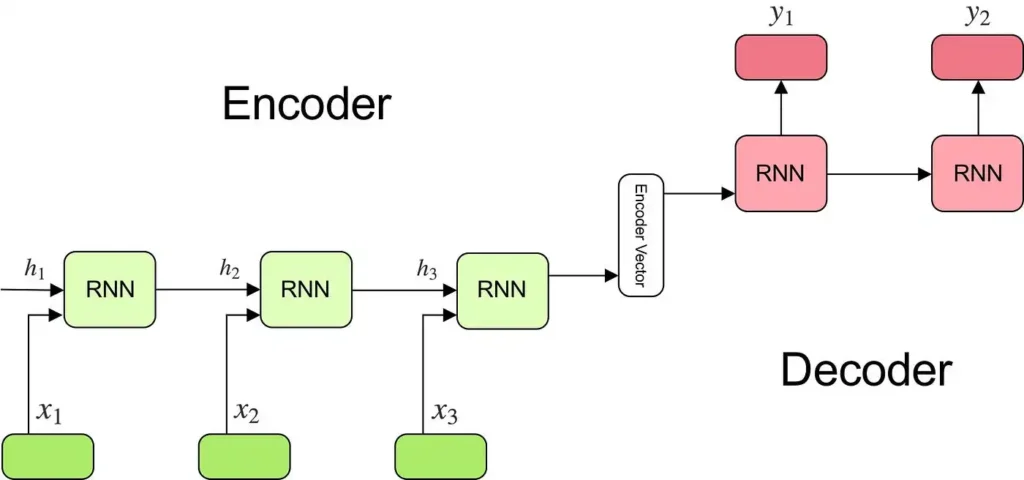
Biology in Action: These models are widely used in metabolic pathway predictions, where genetic data is transformed into insights about biochemical processes.
How Encoder-Decoder Models Work: During encoding, the model captures essential information and discards irrelevant details. In decoding, it uses this compact representation to generate the output. For example, an encoder might simplify a complex cell image, and the decoder would translate it into a disease classification.
6. Autoencoders: Simplifying Complex Data
Biological datasets, such as those from single-cell sequencing, are often high-dimensional and challenging to analyze. Autoencoders help simplify this complexity by reducing data into smaller, more manageable representations without losing important information.

- How Autoencoders Work: Similar to encoder-decoder models, autoencoders compress and reconstruct data. However, their primary goal is to learn efficient representations, which are useful for tasks like anomaly detection or noise reduction.
- Biology in Action: Autoencoders are used to identify rare cell types in single-cell RNA sequencing data or to denoise noisy microscopy images, making them clearer for further analysis.
7. Graph Neural Networks (GNNs): AI for Relationships
Many biological systems, such as protein-protein interaction networks or ecological food webs, are naturally represented as graphs. GNNs are designed to analyze these relationships by understanding how nodes (e.g., proteins or species) connect through edges (e.g., interactions or dependencies).
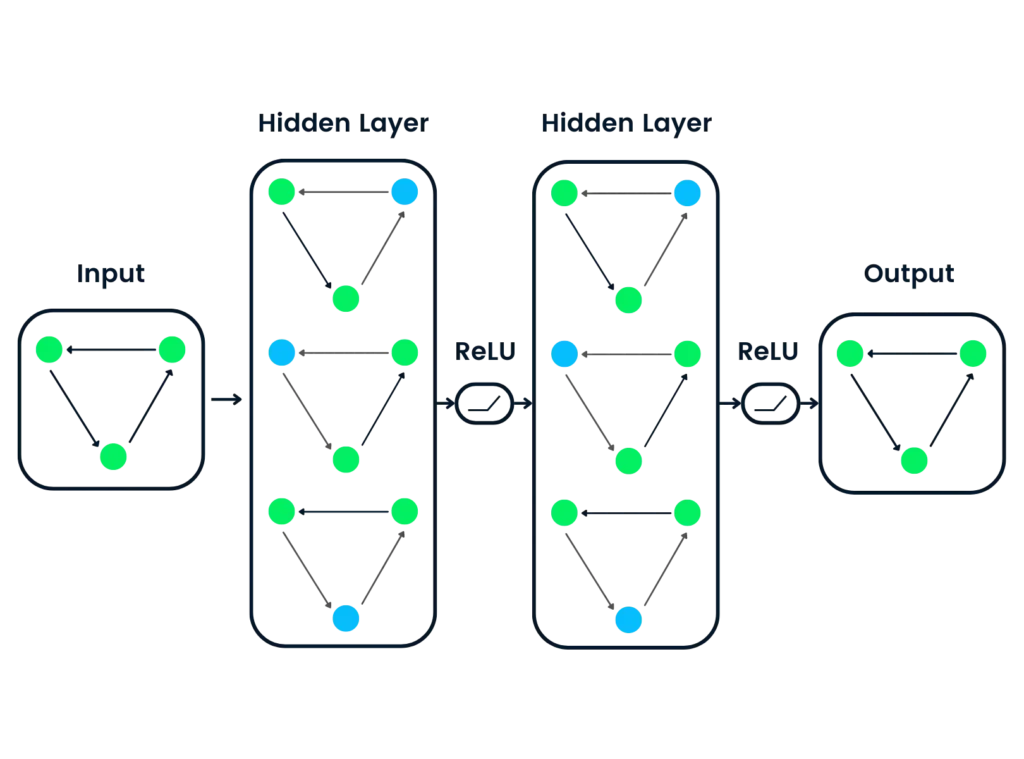
Biology in Action: GNNs are used to predict interactions in protein networks, helping identify potential therapeutic targets. They’re also applied to ecological studies, modeling how species in an ecosystem influence one another.
How GNNs Work: GNNs process both the features of individual nodes and the connections between them. For example, in a drug-target interaction graph, the model evaluates not just the properties of a drug and its target but also their surrounding network of relationships.
8. Reinforcement Learning: AI for Decision-Making
Reinforcement learning teaches models to make decisions by rewarding desirable actions and penalizing undesirable ones. It’s particularly useful for optimizing processes, such as designing experiments or planning therapies.
- How Reinforcement Learning Works: The model starts with no knowledge and learns through trial and error. For instance, in optimizing an experimental protocol, the model tests different conditions and adjusts its strategy based on the outcomes.
- Biology in Action: Reinforcement learning is being used to automate lab workflows, such as optimizing the parameters for CRISPR gene-editing experiments. It’s also applied in adaptive therapies, where treatments are adjusted in real time based on patient responses.
Why These Architectures Matter
Each of these architectures is tailored to specific types of data and problems, making them invaluable tools for biologists. By understanding how they work and where they apply, researchers can unlock new insights and accelerate discoveries in fields ranging from genomics to drug development. Deep learning isn’t just for computer scientists anymore—it’s a powerful ally for advancing life sciences.
Benefits, Challenges, and Future Directions of Deep Learning Architectures for Biologists
Deep learning architectures have transformed how biologists analyze data and make discoveries. From improving efficiency to uncovering hidden patterns in complex datasets, these tools offer significant advantages. However, like any technology, they come with challenges that must be addressed. Understanding these benefits, challenges, and the future of deep learning helps biologists leverage its full potential.
Benefits: Unlocking New Possibilities in Biology
- Faster Analysis of Complex Data
Deep learning accelerates tasks that would take humans months or years to complete. For instance, CNNs can analyze thousands of histopathology slides in hours, helping detect cancer earlier. Similarly, transformers speed up protein structure prediction, a task that once required extensive computational resources and time. - Enhanced Precision and Accuracy
Deep learning architectures excel at identifying subtle patterns in data that traditional methods often miss. For example, autoencoders can detect rare gene expression profiles, while GNNs model intricate protein interaction networks with high accuracy. - Scalability Across Diverse Applications
Whether it’s analyzing genetic sequences, predicting ecological dynamics, or simulating cellular behavior, deep learning architectures adapt to various biological problems. Their flexibility allows biologists to tackle challenges in genomics, drug discovery, and beyond. - Automation of Tedious Tasks
By automating repetitive tasks, such as annotating images or processing sequencing data, deep learning frees researchers to focus on higher-level analysis and hypothesis generation.
Challenges: Navigating the Complexities of AI in Biology
- Data Requirements and Quality
Deep learning models require large, high-quality datasets for training, but biological data is often limited, noisy, or incomplete. For example, rare diseases may lack sufficient data to train effective models, and variability in sample preparation can introduce biases. - Computational Demands
Training deep learning architectures, especially complex ones like transformers or GANs, requires significant computational resources. This can be a barrier for smaller labs or organizations with limited access to high-performance computing. - Interpretability and Trust
Deep learning models are often criticized for being “black boxes,” meaning their decision-making processes are difficult to understand. In fields like biology and medicine, where decisions impact lives, this lack of interpretability can hinder trust and adoption. - Integration with Domain Knowledge
While deep learning is powerful, it sometimes lacks the ability to incorporate biological insights directly. For example, without integrating prior knowledge about biochemical pathways, a model may miss biologically meaningful patterns.
Future Directions: Paving the Way for New Innovations
Emerging Architectures Beyond Deep Learning
As AI evolves, new architectures may surpass current deep learning methods. For instance, neural architecture search (NAS) automates the design of optimal networks, while neuromorphic computing explores brain-inspired hardware to improve efficiency.
Improved Data Utilization
Techniques like data augmentation, transfer learning, and synthetic data generation (using GANs) will help overcome data limitations. These methods allow models to learn effectively from smaller or incomplete datasets, making deep learning more accessible to biologists.
Hybrid Models for Greater Accuracy
Combining deep learning architectures with domain-specific approaches will enhance their performance. For instance, hybrid models integrating GNNs and transformers can better predict complex interactions in protein networks.
Explainable AI (XAI) in Biology
Research into explainable AI will make models more transparent, helping researchers understand why a model predicts certain outcomes. This will increase trust in deep learning applications, especially in high-stakes areas like drug development and diagnostics.
Democratization of AI Tools
Advances in cloud computing and pre-trained models will make deep learning tools more accessible to smaller labs and individual researchers. Biologists will increasingly have access to user-friendly platforms for deploying AI in their research.
FAQs: Common Questions About Deep Learning Architectures for Biologists
- What is a deep learning architecture, and why should biologists care?
Deep learning architectures are specialized AI models designed to analyze complex data. Biologists can use them to process images, analyze genetic sequences, predict protein structures, and discover new drugs—tasks that are difficult or time-consuming with traditional methods. - How is deep learning different from traditional statistical methods?
Traditional methods often rely on pre-defined rules or assumptions, while deep learning automatically learns patterns from data. This makes deep learning particularly effective for handling large, unstructured datasets like genomic sequences or medical images. - Which deep learning architecture is best for biology?
The best architecture depends on the task. For image analysis, CNNs are highly effective. RNNs are ideal for sequential data, like DNA or protein sequences. Transformers and GNNs are powerful for modeling complex relationships in genomic and molecular data. - Do I need programming skills to use deep learning in my research?
While programming skills (e.g., Python) can help, many tools and platforms now offer user-friendly interfaces for deploying deep learning models. Researchers can use pre-trained models or collaborate with data scientists for more complex tasks. - What are the limitations of deep learning in biology?
Key challenges include the need for large, high-quality datasets, the computational resources required for training models, and the difficulty in interpreting results. However, advances like synthetic data generation and explainable AI are helping address these issues.
Conclusion: Deep Learning as a Catalyst for Biological Discovery
Deep learning architectures are transforming biology, offering tools that accelerate discoveries, enhance precision, and open doors to previously unattainable insights. From CNNs analyzing complex images to transformers predicting protein structures, these AI models empower researchers to tackle biological challenges with unprecedented efficiency.
However, their adoption comes with challenges, including data limitations, computational demands, and interpretability issues. Addressing these hurdles will require collaboration between biologists, data scientists, and AI developers. The future holds even greater promise, with innovations like hybrid models, explainable AI, and accessible tools making deep learning more powerful and easier to use.
Whether you’re analyzing genetic sequences, designing new drugs, or exploring ecological networks, deep learning architectures are an essential part of the biologist’s toolkit. By embracing these technologies, researchers can push the boundaries of life sciences, solving problems faster and uncovering breakthroughs that benefit both science and society.
References
- Alzubaidi, L., Zhang, J., Humaidi, A.J. et al. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. J Big Data 8, 53 (2021). https://doi.org/10.1186/s40537-021-00444-8
- Sarker, I.H. Deep Learning: A Comprehensive Overview on Techniques, Taxonomy, Applications and Research Directions. SN COMPUT. SCI. 2, 420 (2021). https://doi.org/10.1007/s42979-021-00815-1